Call closure_generate() to run the CLOSURE algorithm on a
given set of summary statistics.
This can take seconds, minutes, or longer, depending on the input. Wide
variance and large n often lead to many samples, i.e., long runtimes.
These effects interact dynamically. For example, with large n, even very
small increases in sd can greatly increase runtime and number of values
found.
If the inputs are inconsistent, there is no solution. The function will then return empty results and throw a warning.
Usage
closure_generate(
mean,
sd,
n,
scale_min,
scale_max,
rounding = "up_or_down",
threshold = 5,
warn_if_empty = TRUE,
ask_to_proceed = TRUE,
rounding_error_mean = NULL,
rounding_error_sd = NULL
)Arguments
- mean
String (length 1). Reported mean.
- sd
String (length 1). Reported sample standard deviation.
- n
Numeric (length 1). Reported sample size.
- scale_min, scale_max
Numeric (length 1 each). Minimal and maximal possible values of the measurement scale. For example, with a 1-7 Likert scale, use
scale_min = 1andscale_max = 7. Prefer the empirical min and max if available: they constrain the possible values further.- rounding
String (length 1). Rounding method assumed to have created
meanandsd. See Rounding options, but also the Rounding limitations section below. Default is"up_or_down"which, e.g., unrounds0.12to0.115as a lower bound and0.125as an upper bound.- threshold
Numeric (length 1). Number from which to round up or down, if
roundingis any of"up_or_down","up", and"down". Default is5.- warn_if_empty
Logical (length 1). Should a warning be shown if no samples are found? Default is
TRUE.- ask_to_proceed
Logical (length 1). If the runtime is predicted to be very long, should the function prompt you to proceed or abort in an interactive setting? Default is
TRUE.- rounding_error_mean, rounding_error_sd
Numeric (length 1 each). Option to manually set the rounding error around
meanandsd. This is meant for development and might be removed in the future, so most users can ignore it.
Value
Named list of four tibbles (data frames):
inputs: Arguments to this function.metrics:samples_initial: integer. The basis for computing CLOSURE results, based on scale range only. Seeclosure_count_initial().samples_all: integer. Number of all samples. Equal to the number of rows inresults.values_all: integer. Number of all individual values found. Equal ton * samples_all.horns: double. Measure of dispersion for bounded scales; seehorns().horns_uniform: double. Valuehornswould have if the reconstructed sample was uniformly distributed.
frequency:value: integer. Scale values derived fromscale_minandscale_max.f_average: Count of scale values in the meanresultssample.f_absolute: integer. Count of individual scale values found in theresultssamples.f_relative: double. Values' share of total values found.
results:id: integer. Runs from1tosamples_all.sample: list of integer vectors. Each of these vectors has lengthn. It is a sample (or distribution) of individual scale values found by CLOSURE.
Rounding limitations
The rounding and threshold arguments are
not fully implemented. For example, CLOSURE currently treats all rounding
bounds as inclusive, even if the rounding specification would imply
otherwise.
Many specifications of the two arguments will not make any difference, and those that do will most likely lead to empty results.
Examples
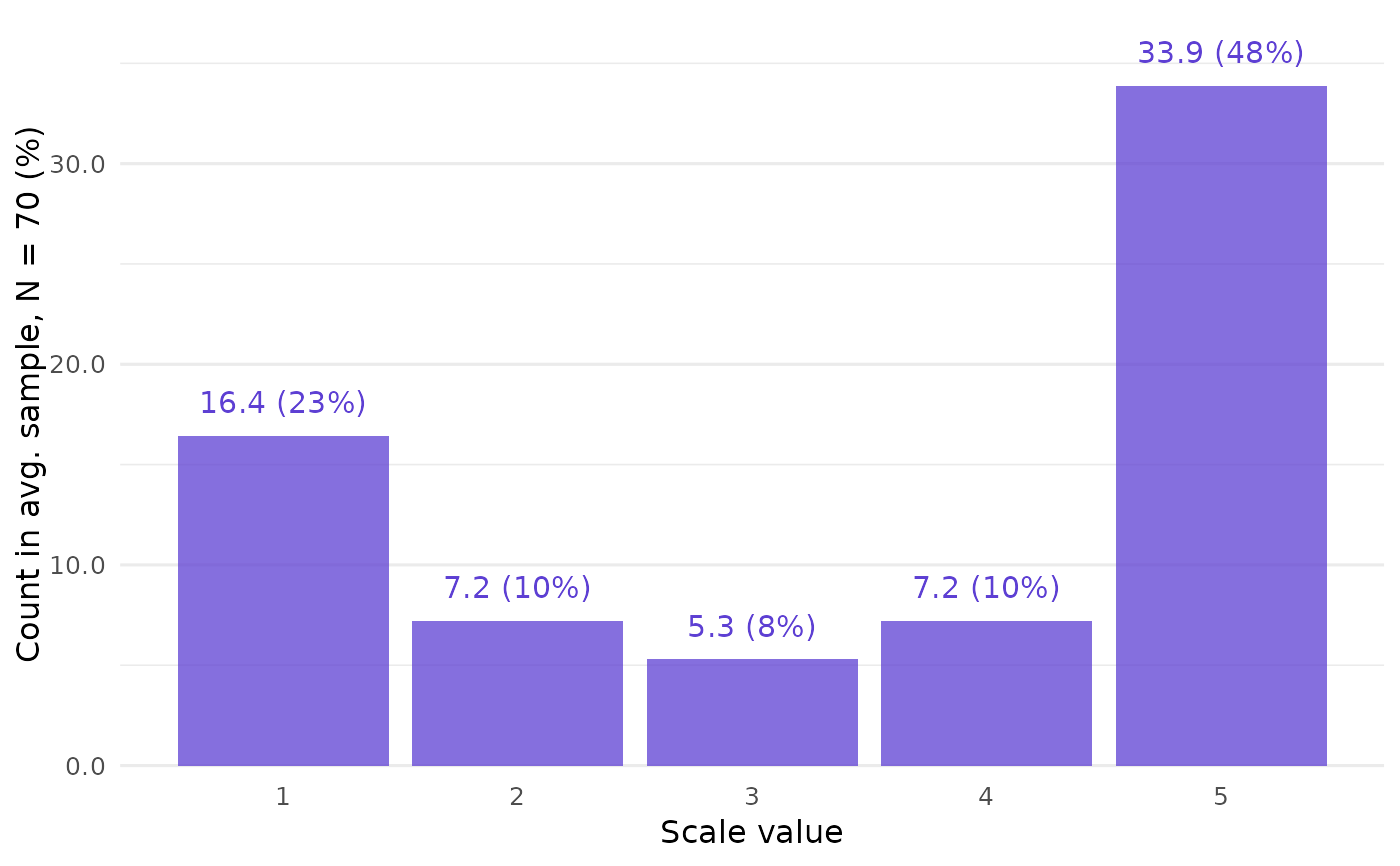
# High spread often leads to many samples --
# here, 3682.
data_high <- closure_generate(
mean = "3.5",
sd = "1.7",
n = 70,
scale_min = 1,
scale_max = 5
)
data_high
#> $inputs
#> # A tibble: 1 × 7
#> mean sd n scale_min scale_max rounding threshold
#> <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl>
#> 1 3.5 1.7 70 1 5 up_or_down 5
#>
#> $metrics
#> # A tibble: 1 × 5
#> samples_initial samples_all values_all horns horns_uniform
#> <int> <int> <int> <dbl> <dbl>
#> 1 15 2492 174440 0.708 0.5
#>
#> $frequency
#> # A tibble: 5 × 4
#> value f_average f_absolute f_relative
#> <int> <dbl> <int> <dbl>
#> 1 1 16.4 40982 0.235
#> 2 2 7.19 17922 0.103
#> 3 3 5.29 13172 0.0755
#> 4 4 7.19 17916 0.103
#> 5 5 33.9 84448 0.484
#>
#> $results
#> # A tibble: 2,492 × 2
#> id sample
#> <int> <list>
#> 1 1 <int [70]>
#> 2 2 <int [70]>
#> 3 3 <int [70]>
#> 4 4 <int [70]>
#> 5 5 <int [70]>
#> 6 6 <int [70]>
#> 7 7 <int [70]>
#> 8 8 <int [70]>
#> 9 9 <int [70]>
#> 10 10 <int [70]>
#> # ℹ 2,482 more rows
#>
# Get a clear picture of the distribution
# by following up with `closure_plot_bar()`:
closure_plot_bar(data_high)
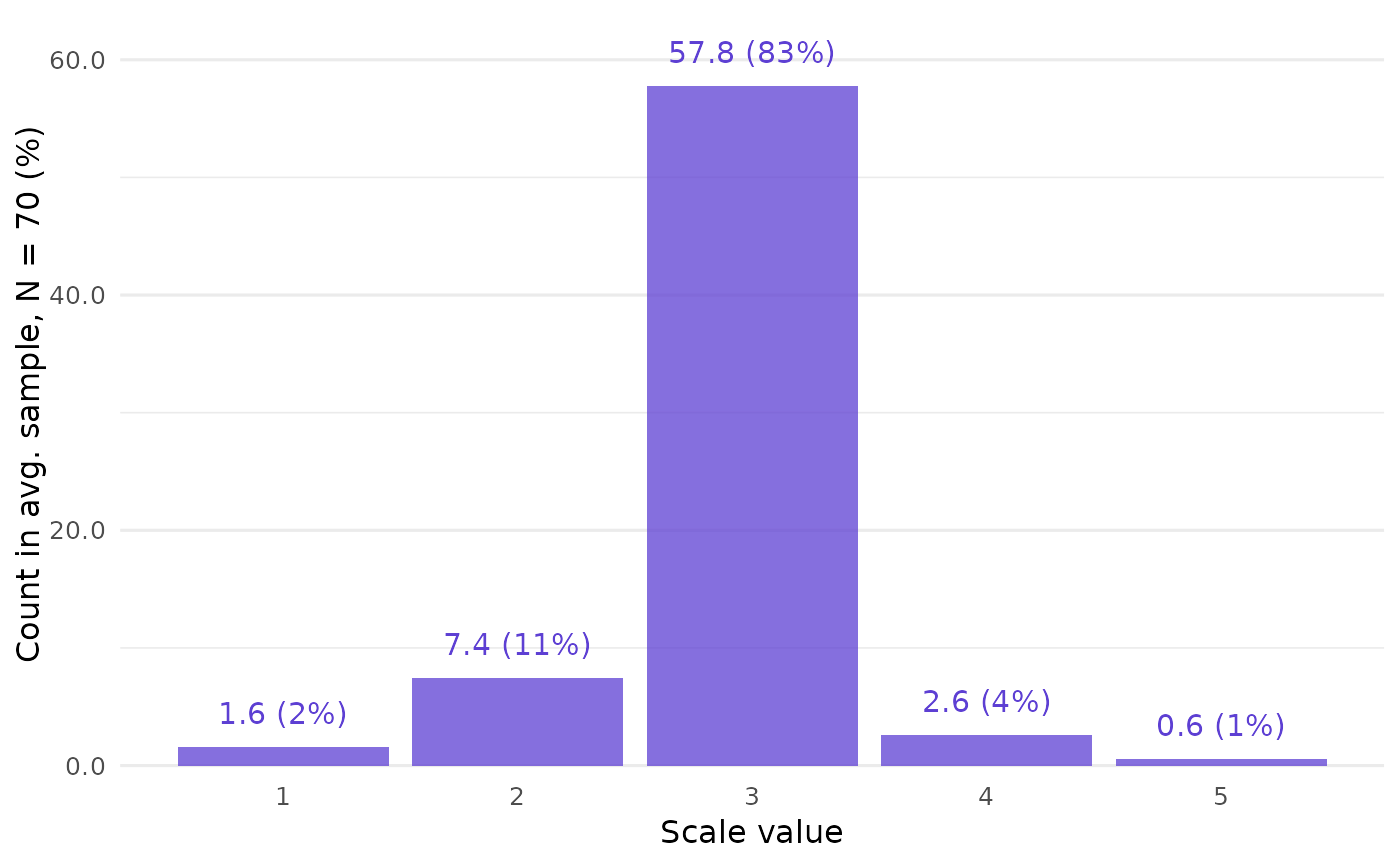
 # Low spread, only 3 samples, and not all
# scale values are possible.
data_low <- closure_generate(
mean = "2.9",
sd = "0.5",
n = 70,
scale_min = 1,
scale_max = 5
)
data_low
#> $inputs
#> # A tibble: 1 × 7
#> mean sd n scale_min scale_max rounding threshold
#> <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl>
#> 1 2.9 0.5 70 1 5 up_or_down 5
#>
#> $metrics
#> # A tibble: 1 × 5
#> samples_initial samples_all values_all horns horns_uniform
#> <int> <int> <int> <dbl> <dbl>
#> 1 15 219 15330 0.0643 0.5
#>
#> $frequency
#> # A tibble: 5 × 4
#> value f_average f_absolute f_relative
#> <int> <dbl> <int> <dbl>
#> 1 1 1.59 349 0.0228
#> 2 2 7.42 1626 0.106
#> 3 3 57.8 12659 0.826
#> 4 4 2.61 572 0.0373
#> 5 5 0.566 124 0.00809
#>
#> $results
#> # A tibble: 219 × 2
#> id sample
#> <int> <list>
#> 1 1 <int [70]>
#> 2 2 <int [70]>
#> 3 3 <int [70]>
#> 4 4 <int [70]>
#> 5 5 <int [70]>
#> 6 6 <int [70]>
#> 7 7 <int [70]>
#> 8 8 <int [70]>
#> 9 9 <int [70]>
#> 10 10 <int [70]>
#> # ℹ 209 more rows
#>
# This can also be shown by `closure_plot_bar()`:
closure_plot_bar(data_low)
# Low spread, only 3 samples, and not all
# scale values are possible.
data_low <- closure_generate(
mean = "2.9",
sd = "0.5",
n = 70,
scale_min = 1,
scale_max = 5
)
data_low
#> $inputs
#> # A tibble: 1 × 7
#> mean sd n scale_min scale_max rounding threshold
#> <chr> <chr> <dbl> <dbl> <dbl> <chr> <dbl>
#> 1 2.9 0.5 70 1 5 up_or_down 5
#>
#> $metrics
#> # A tibble: 1 × 5
#> samples_initial samples_all values_all horns horns_uniform
#> <int> <int> <int> <dbl> <dbl>
#> 1 15 219 15330 0.0643 0.5
#>
#> $frequency
#> # A tibble: 5 × 4
#> value f_average f_absolute f_relative
#> <int> <dbl> <int> <dbl>
#> 1 1 1.59 349 0.0228
#> 2 2 7.42 1626 0.106
#> 3 3 57.8 12659 0.826
#> 4 4 2.61 572 0.0373
#> 5 5 0.566 124 0.00809
#>
#> $results
#> # A tibble: 219 × 2
#> id sample
#> <int> <list>
#> 1 1 <int [70]>
#> 2 2 <int [70]>
#> 3 3 <int [70]>
#> 4 4 <int [70]>
#> 5 5 <int [70]>
#> 6 6 <int [70]>
#> 7 7 <int [70]>
#> 8 8 <int [70]>
#> 9 9 <int [70]>
#> 10 10 <int [70]>
#> # ℹ 209 more rows
#>
# This can also be shown by `closure_plot_bar()`:
closure_plot_bar(data_low)